Articles in press have been peer-reviewed and accepted, which are not yet assigned to volumes /issues, but are citable by Digital Object Identifier (DOI).
Column
2023, 9(6): 299-308.
doi: 10.52601/bpr.2023.230042
Abstract:
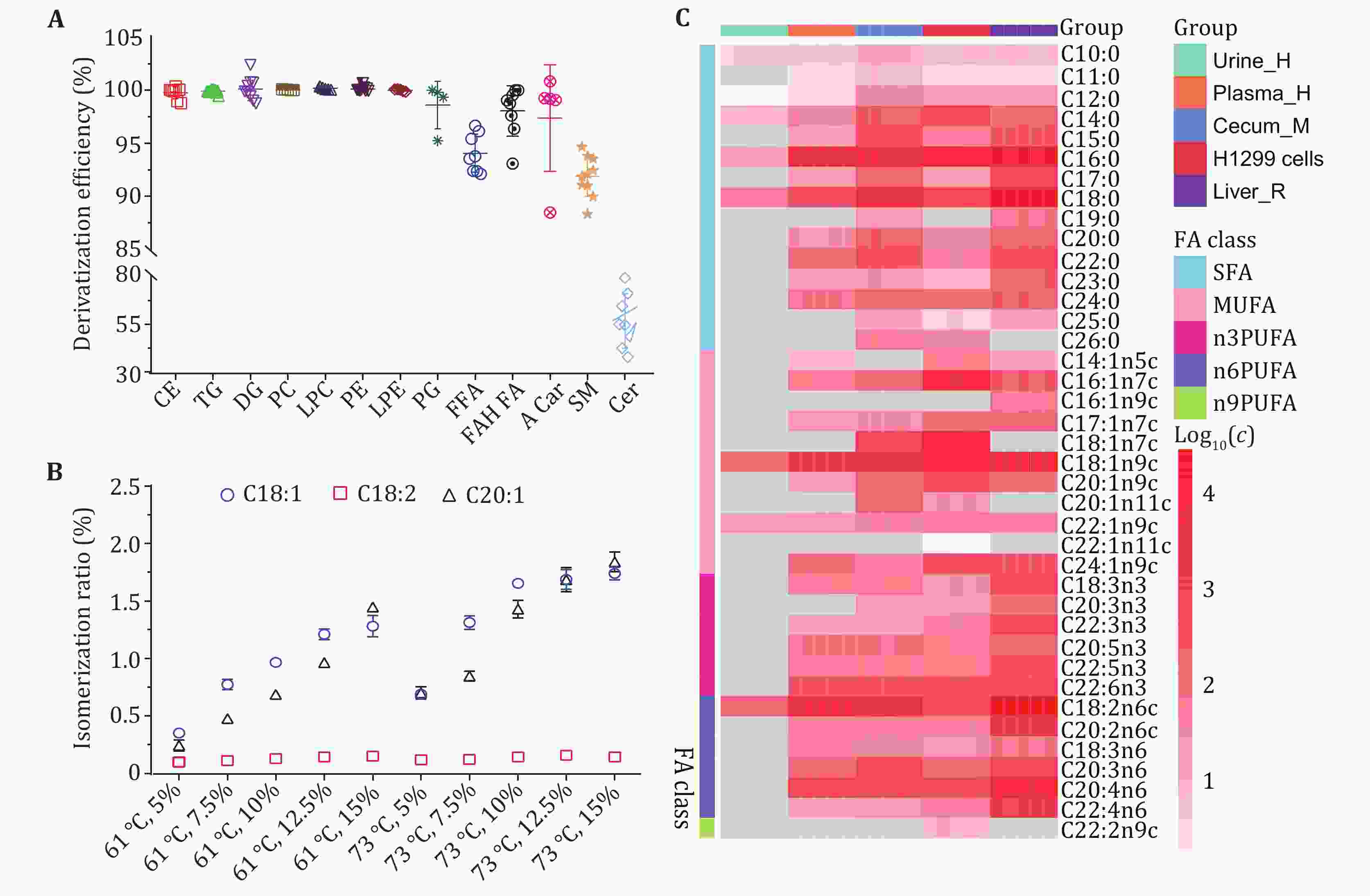
Efficient quantification of fatty-acid (FA) composition (fatty-acidome) in biological samples is crucial for understanding physiology and pathophysiology in large population cohorts. Here, we report a rapid GC-FID/MS method for simultaneous quantification of all FAs in numerous biological matrices. Within eight minutes, this method enabled simultaneous quantification of 50 FAs as fatty-acid methyl esters (FAMEs) in femtomole levels following the efficient transformation of FAs in all lipids including FFAs, cholesterol-esters, glycerides, phospholipids and sphingolipids. The method showed satisfactory inter-day and intra-day precision, stability and linearity (R2 > 0.994) within a concentration range of 2–3 orders of magnitude. FAs were then quantified in typical multiple biological matrices including human biofluids (urine, plasma) and cells, animal intestinal content and tissue samples. We also established a quantitative structure-retention relationship (QSRR) for analytes to accurately predict their retention time and aid their reliable identification. We further developed a novel no-additive retention index (NARI) with endogenous FAMEs reducing inter-batch variations to 15 seconds; such NARI performed better than the alkanes-based classical RI, making meta-analysis possible for data obtained from different batches and platforms. Collectively, this provides an inexpensive high-throughput analytical system for quantitative phenotyping of all FAs in 8-minutes multiple biological matrices in large cohort studies of pathophysiological effects.
Efficient quantification of fatty-acid (FA) composition (fatty-acidome) in biological samples is crucial for understanding physiology and pathophysiology in large population cohorts. Here, we report a rapid GC-FID/MS method for simultaneous quantification of all FAs in numerous biological matrices. Within eight minutes, this method enabled simultaneous quantification of 50 FAs as fatty-acid methyl esters (FAMEs) in femtomole levels following the efficient transformation of FAs in all lipids including FFAs, cholesterol-esters, glycerides, phospholipids and sphingolipids. The method showed satisfactory inter-day and intra-day precision, stability and linearity (R2 > 0.994) within a concentration range of 2–3 orders of magnitude. FAs were then quantified in typical multiple biological matrices including human biofluids (urine, plasma) and cells, animal intestinal content and tissue samples. We also established a quantitative structure-retention relationship (QSRR) for analytes to accurately predict their retention time and aid their reliable identification. We further developed a novel no-additive retention index (NARI) with endogenous FAMEs reducing inter-batch variations to 15 seconds; such NARI performed better than the alkanes-based classical RI, making meta-analysis possible for data obtained from different batches and platforms. Collectively, this provides an inexpensive high-throughput analytical system for quantitative phenotyping of all FAs in 8-minutes multiple biological matrices in large cohort studies of pathophysiological effects.
2023, 9(6): 309-324.
doi: 10.52601/bpr.2023.230034
Abstract:
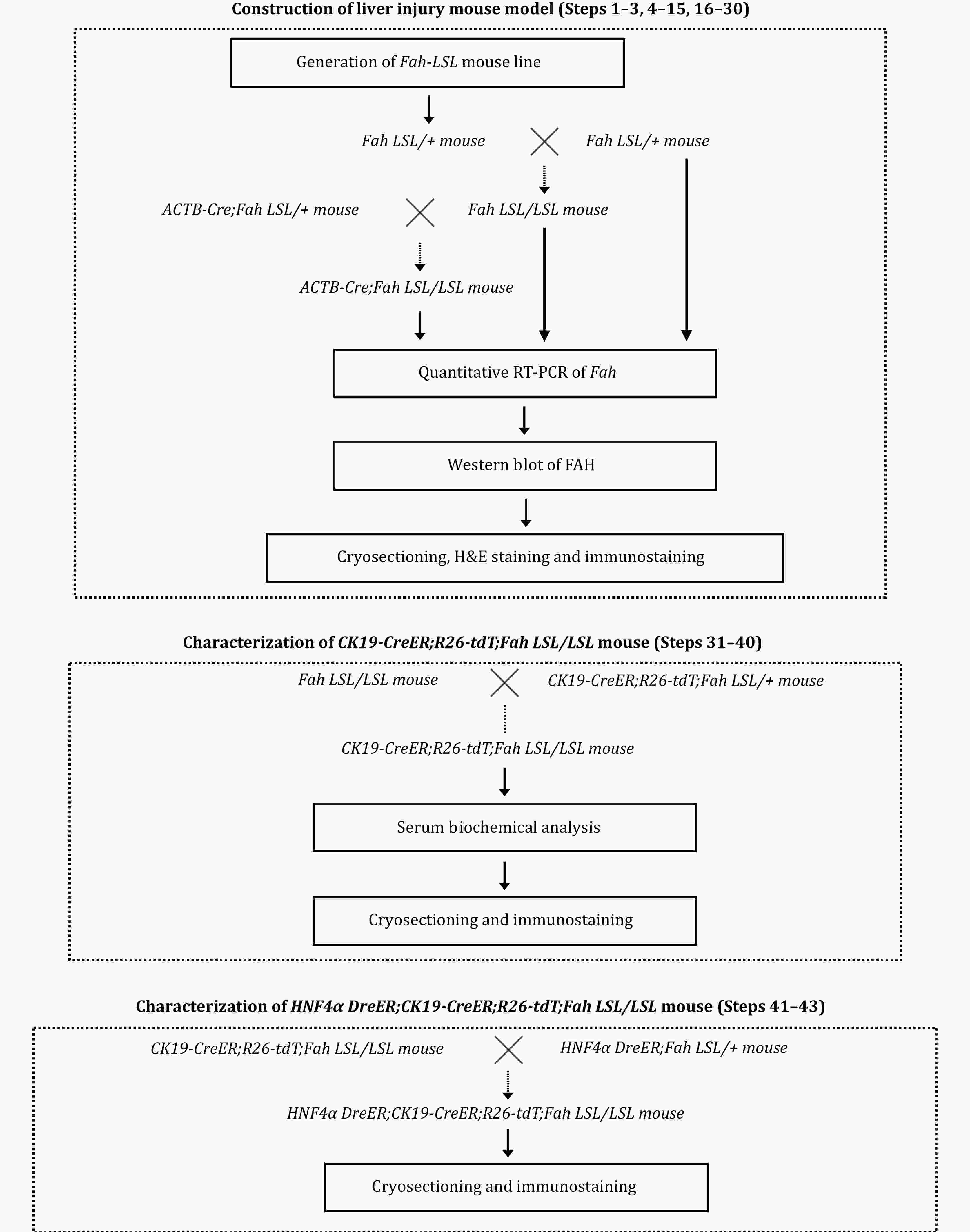
The liver consists predominantly of hepatocytes and biliary epithelial cells (BECs), which serve distinct physiological functions. Although hepatocytes primarily replenish their own population during homeostasis and injury repair, recent findings have suggested that BECs can transdifferentiate into hepatocytes when hepatocyte-mediated liver regeneration is impaired. However, the cellular and molecular mechanisms governing this BEC-to-hepatocyte conversion remain poorly understood largely because of the inefficiency of existing methods for inducing lineage conversion. Therefore, this study introduces a novel mouse model engineered by the Zhou's lab, where hepatocyte senescence is induced by the deletion of the fumarylacetoacetate (Fah) gene. This model facilitates the efficient conversion of BECs to hepatocytes and allows for the simultaneous lineage tracing of BECs; consequently, a transitional liver progenitor cell population can be identified during lineage conversion. This study also outlines the technical procedures for utilizing this model to determine the underlying cellular and molecular mechanisms of BEC-to-hepatocyte conversion and provides new insights into liver regeneration and its underlying molecular mechanism.
The liver consists predominantly of hepatocytes and biliary epithelial cells (BECs), which serve distinct physiological functions. Although hepatocytes primarily replenish their own population during homeostasis and injury repair, recent findings have suggested that BECs can transdifferentiate into hepatocytes when hepatocyte-mediated liver regeneration is impaired. However, the cellular and molecular mechanisms governing this BEC-to-hepatocyte conversion remain poorly understood largely because of the inefficiency of existing methods for inducing lineage conversion. Therefore, this study introduces a novel mouse model engineered by the Zhou's lab, where hepatocyte senescence is induced by the deletion of the fumarylacetoacetate (Fah) gene. This model facilitates the efficient conversion of BECs to hepatocytes and allows for the simultaneous lineage tracing of BECs; consequently, a transitional liver progenitor cell population can be identified during lineage conversion. This study also outlines the technical procedures for utilizing this model to determine the underlying cellular and molecular mechanisms of BEC-to-hepatocyte conversion and provides new insights into liver regeneration and its underlying molecular mechanism.
2023, 9(6): 325-337.
doi: 10.52601/bpr.2023.230029
Abstract:
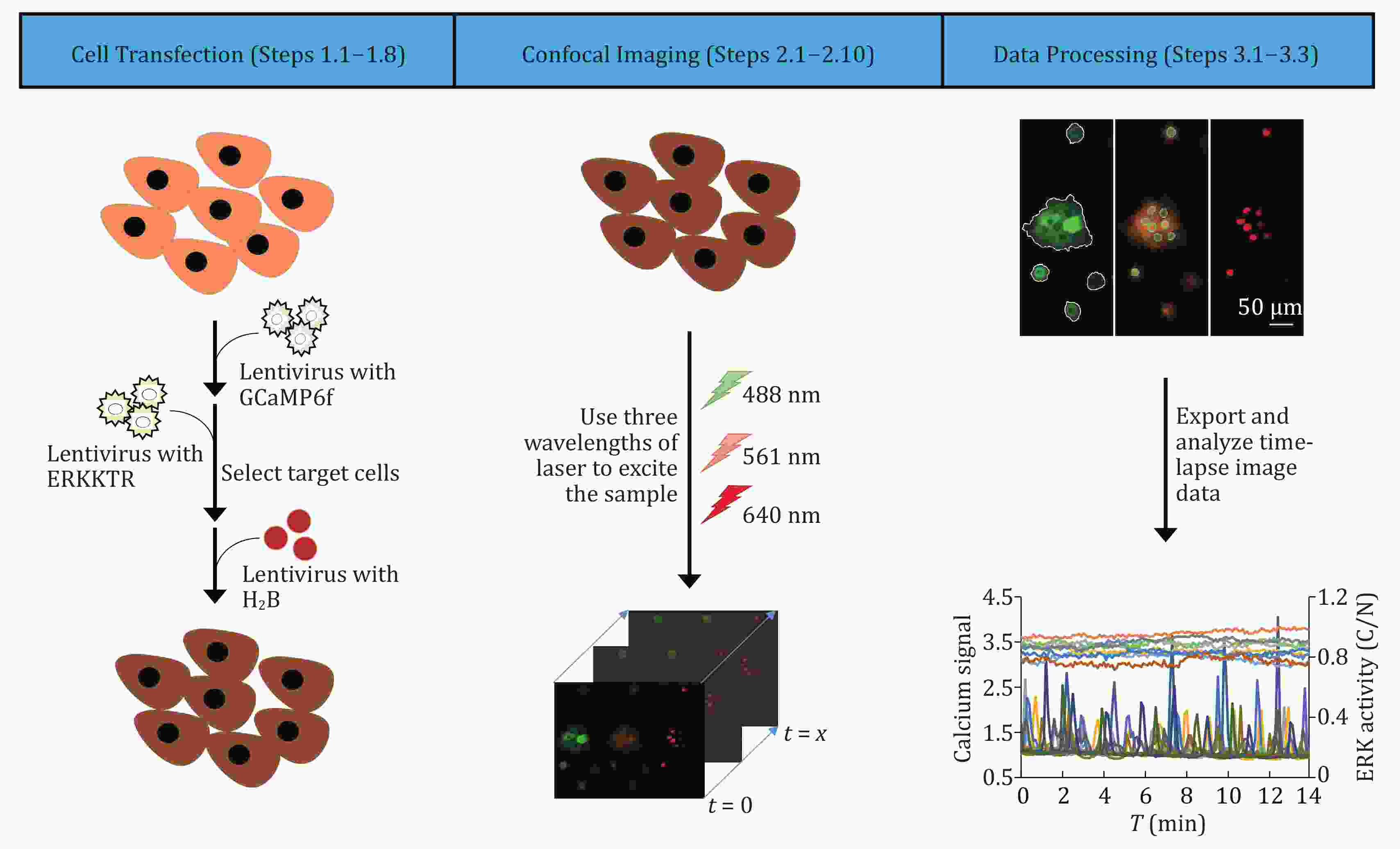
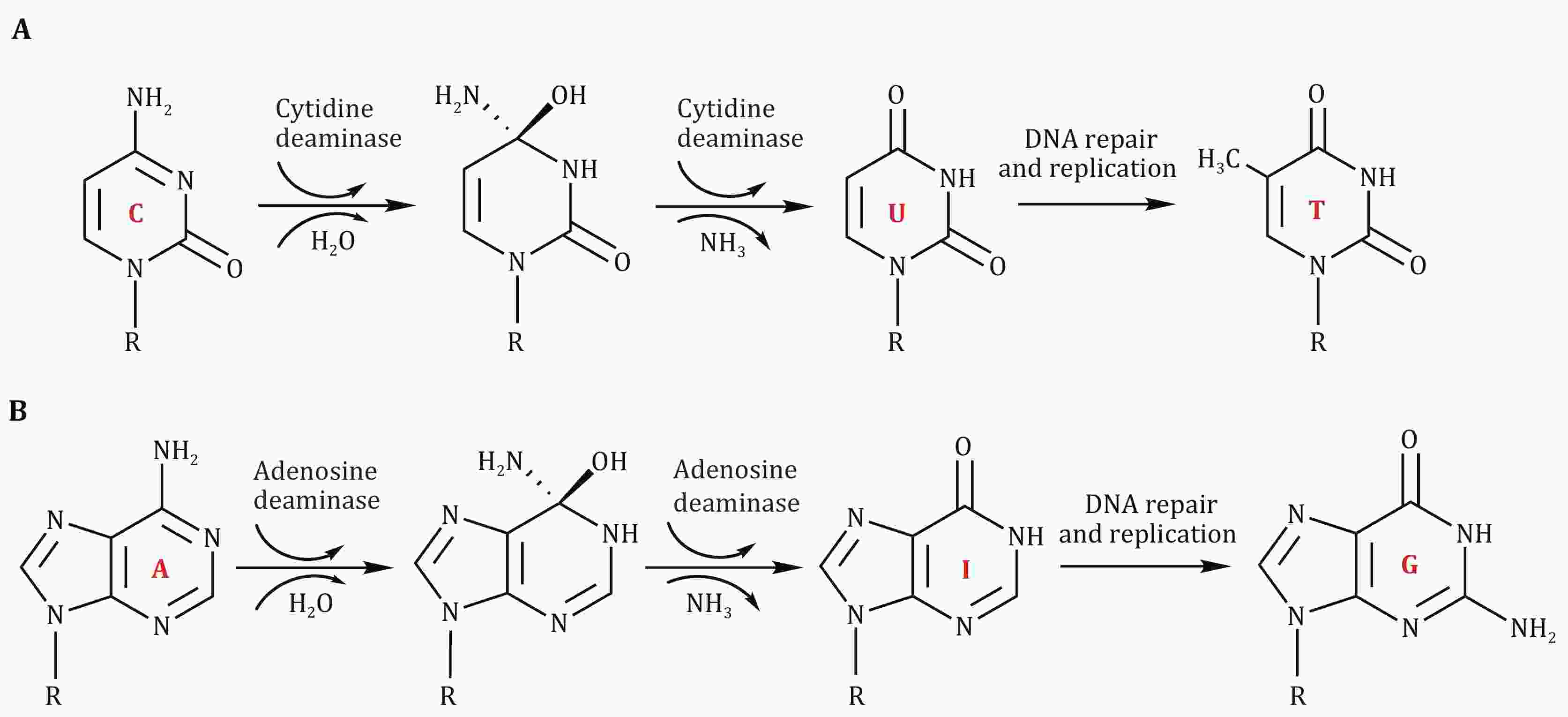
The development of nucleoside deaminase-containing base editors realized targeted single base change with high efficiency and precision. Such nucleoside deaminases include adenosine and cytidine deaminases, which can catalyze adenosine-to-inosine (A-to-I) and cytidine-to-uridine (C-to-U) conversion respectively. These nucleoside deaminases are under the spotlight because of their vast application potential in gene editing. Recent advances in the engineering of current nucleoside deaminases and the discovery of new nucleoside deaminases greatly broaden the application scope and improve the editing specificity of base editors. In this review, we cover current knowledge about the deaminases used in base editors, including their key structural features, working mechanisms, optimization, and evolution.
The development of nucleoside deaminase-containing base editors realized targeted single base change with high efficiency and precision. Such nucleoside deaminases include adenosine and cytidine deaminases, which can catalyze adenosine-to-inosine (A-to-I) and cytidine-to-uridine (C-to-U) conversion respectively. These nucleoside deaminases are under the spotlight because of their vast application potential in gene editing. Recent advances in the engineering of current nucleoside deaminases and the discovery of new nucleoside deaminases greatly broaden the application scope and improve the editing specificity of base editors. In this review, we cover current knowledge about the deaminases used in base editors, including their key structural features, working mechanisms, optimization, and evolution.
2023, 9(6): 338-351.
doi: 10.52601/bpr.2023.230032
Abstract:
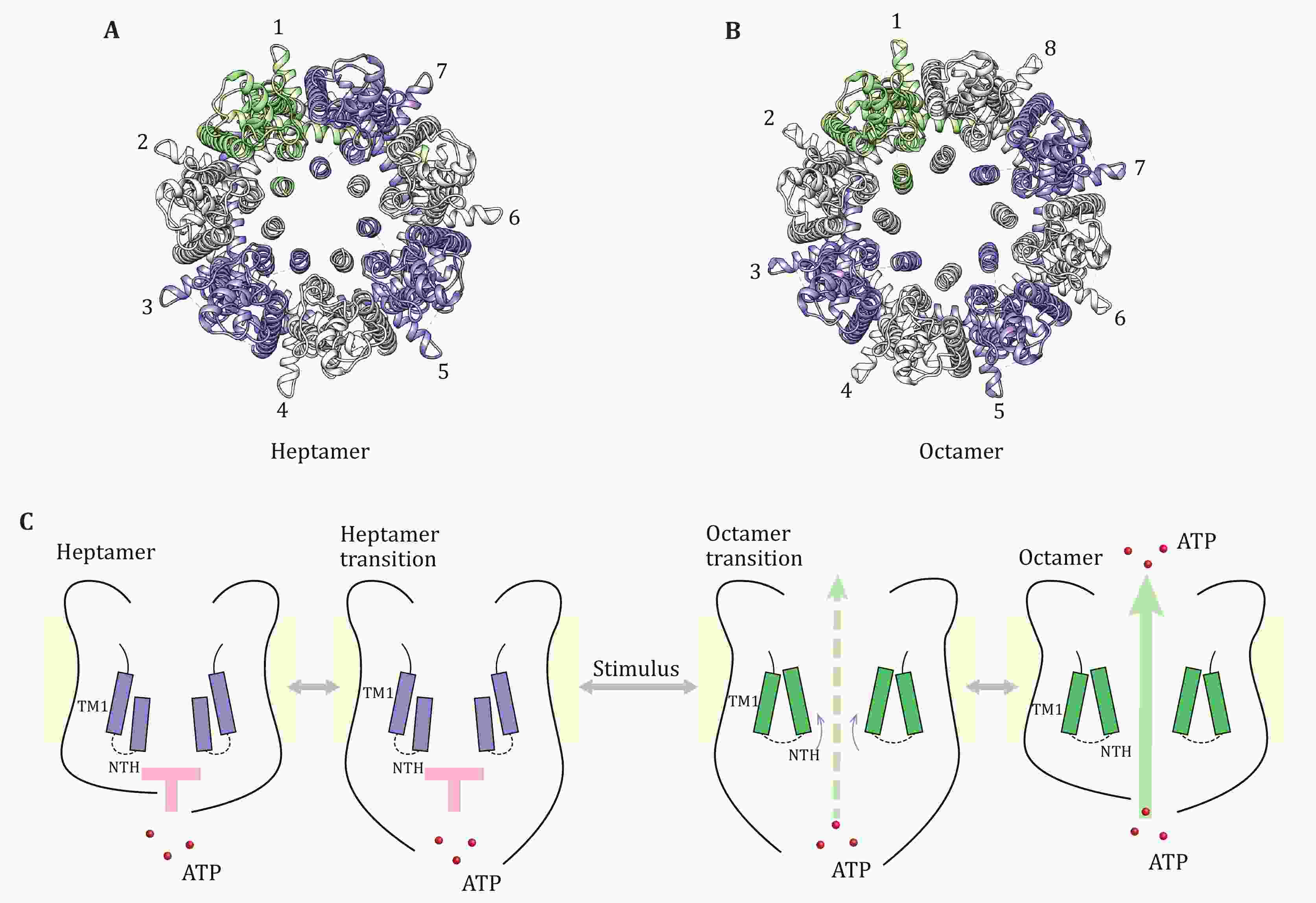
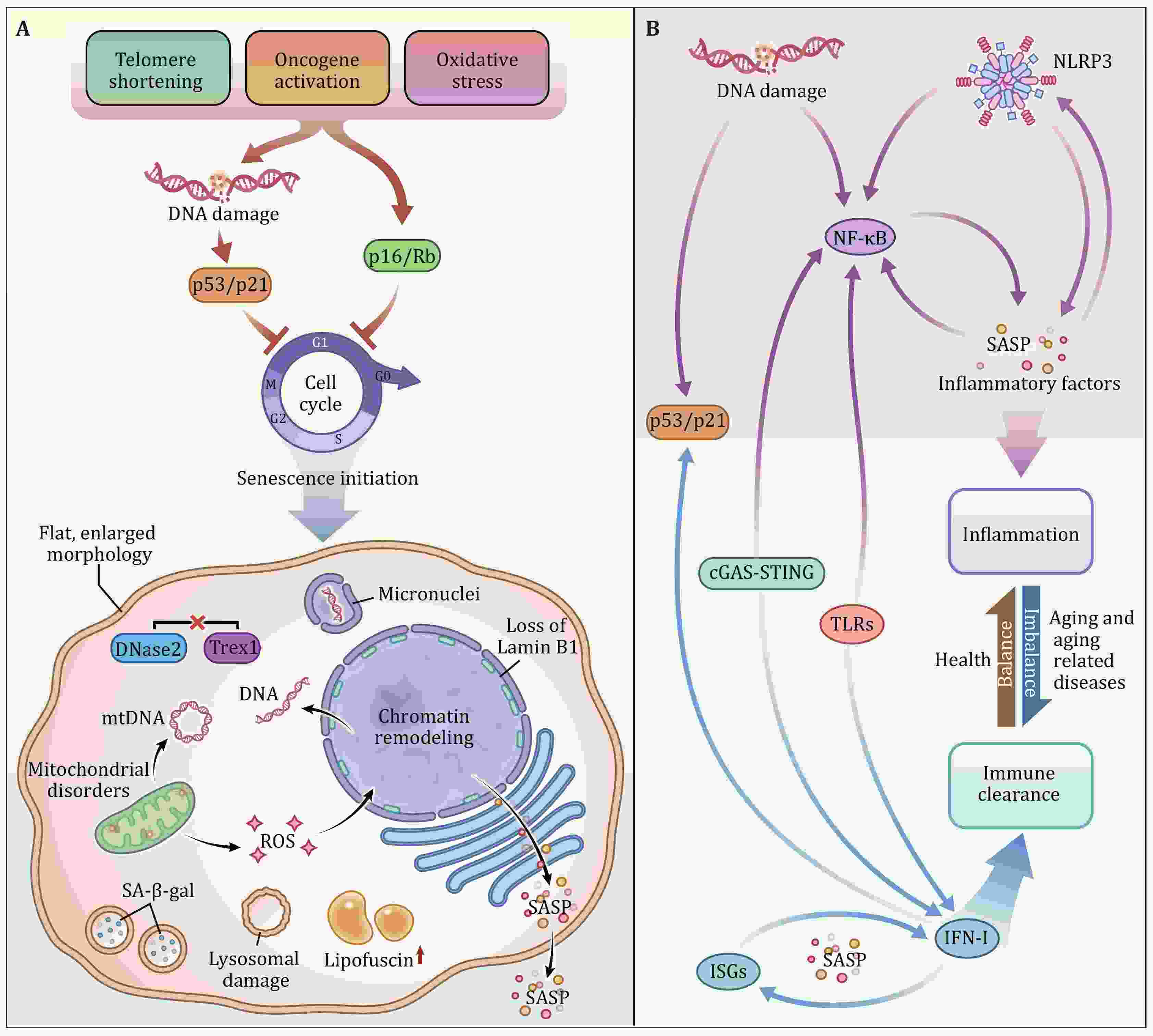
During the COVID-19 pandemic, the interplay between the processes of immunity and senescence is drawing more and more intensive attention. SARS-CoV-2 infection induces senescence in lung cells, failure to clear infected cells and increased presence of inflammatory factors could lead to a cytokine storm and acute respiratory disease syndrome (ARDS), which together with aging and age-associated disease lead to 70% of COVID-19-related deaths. Studies on how senescence initiates upon viral infection and how to restrict excessive accumulation of senescent cells to avoid harmful inflammation are crucially important. Senescence can induce innate immune signaling, and innate immunity can engage cell senescence. Here, we mainly review the innate immune pathways, such as cGAS-STING, TLRs, NF-κB, and NLRP3 inflammasome, participating in the senescence process. In these pathways, IFN-I and inflammatory factors play key roles. At the end of the review, we propose the strategies by which we can improve the immune function and reduce inflammation based on these findings.
During the COVID-19 pandemic, the interplay between the processes of immunity and senescence is drawing more and more intensive attention. SARS-CoV-2 infection induces senescence in lung cells, failure to clear infected cells and increased presence of inflammatory factors could lead to a cytokine storm and acute respiratory disease syndrome (ARDS), which together with aging and age-associated disease lead to 70% of COVID-19-related deaths. Studies on how senescence initiates upon viral infection and how to restrict excessive accumulation of senescent cells to avoid harmful inflammation are crucially important. Senescence can induce innate immune signaling, and innate immunity can engage cell senescence. Here, we mainly review the innate immune pathways, such as cGAS-STING, TLRs, NF-κB, and NLRP3 inflammasome, participating in the senescence process. In these pathways, IFN-I and inflammatory factors play key roles. At the end of the review, we propose the strategies by which we can improve the immune function and reduce inflammation based on these findings.
2023, 9(6): 352-361.
doi: 10.52601/bpr.2023.230035
Abstract:
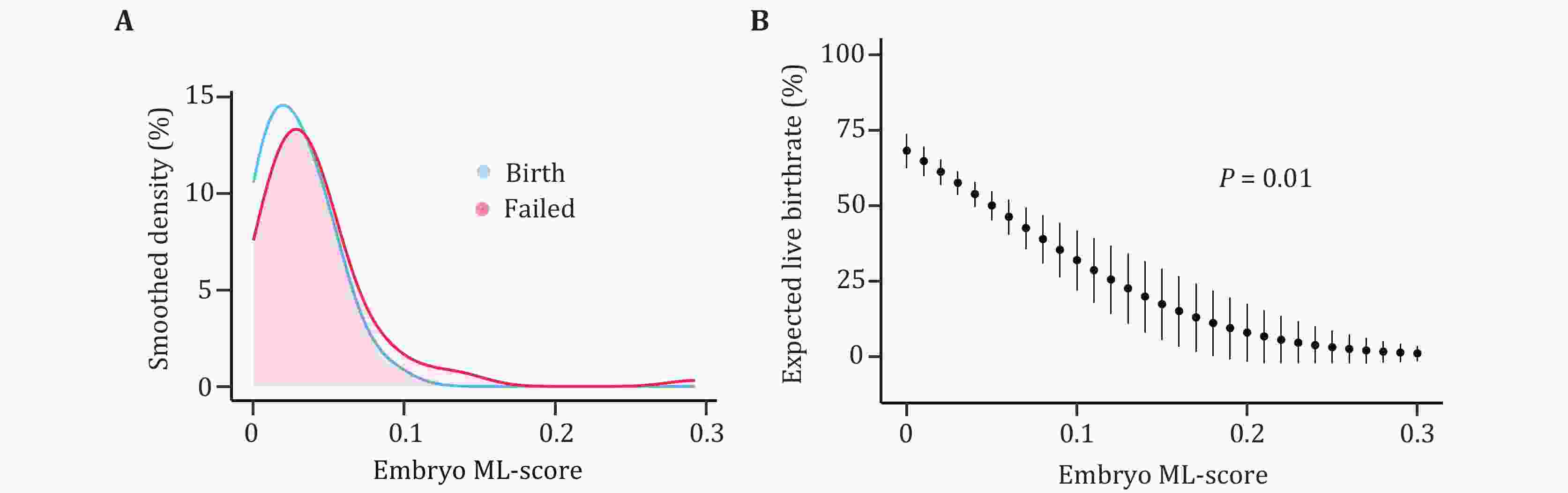
Embryo quality is a critical determinant of clinical outcomes in assisted reproductive technology (ART). A recent clinical trial investigating preimplantation DNA methylation screening (PIMS) revealed that whole genome DNA methylation level is a novel biomarker for assessing ART embryo quality. Here, we reinforced and estimated the clinical efficacy of PIMS. We introduce PIMS-AI, an innovative artificial intelligence (AI) based model, to predict the probability of an embryo producing live birth and subsequently assist ART embryo selection. Our model demonstrated robust performance, achieving an area under the curve (AUC) of 0.90 in cross-validation and 0.80 in independent testing. In simulated embryo selection, PIMS-AI attained an accuracy of 81% in identifying viable embryos for patients. Notably, PIMS-AI offers significant advantages over conventional preimplantation genetic testing for aneuploidy (PGT-A), including enhanced embryo discriminability and the potential to benefit a broader patient population. In conclusion, our approach holds substantial promise for clinical application and has the potential to significantly improve the ART success rate.
Embryo quality is a critical determinant of clinical outcomes in assisted reproductive technology (ART). A recent clinical trial investigating preimplantation DNA methylation screening (PIMS) revealed that whole genome DNA methylation level is a novel biomarker for assessing ART embryo quality. Here, we reinforced and estimated the clinical efficacy of PIMS. We introduce PIMS-AI, an innovative artificial intelligence (AI) based model, to predict the probability of an embryo producing live birth and subsequently assist ART embryo selection. Our model demonstrated robust performance, achieving an area under the curve (AUC) of 0.90 in cross-validation and 0.80 in independent testing. In simulated embryo selection, PIMS-AI attained an accuracy of 81% in identifying viable embryos for patients. Notably, PIMS-AI offers significant advantages over conventional preimplantation genetic testing for aneuploidy (PGT-A), including enhanced embryo discriminability and the potential to benefit a broader patient population. In conclusion, our approach holds substantial promise for clinical application and has the potential to significantly improve the ART success rate.






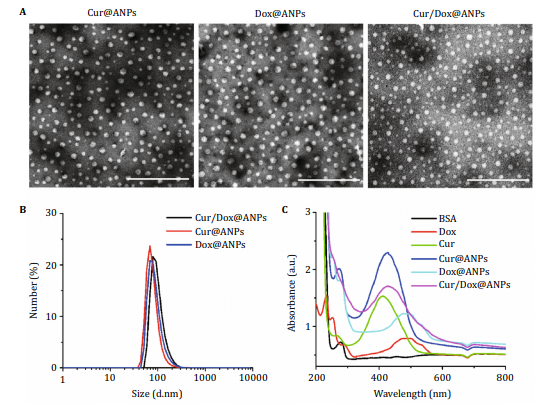
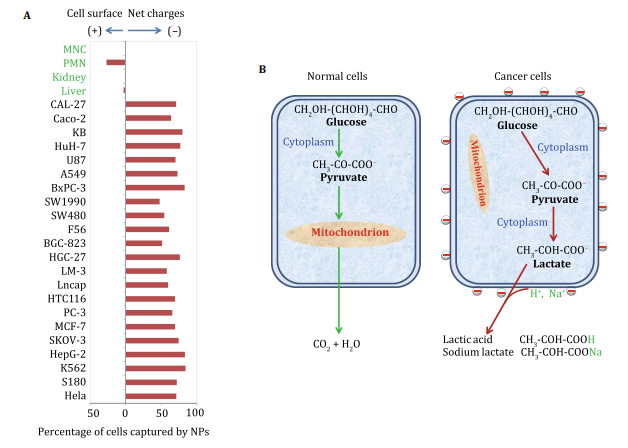



 Abstract
Abstract PDF
PDF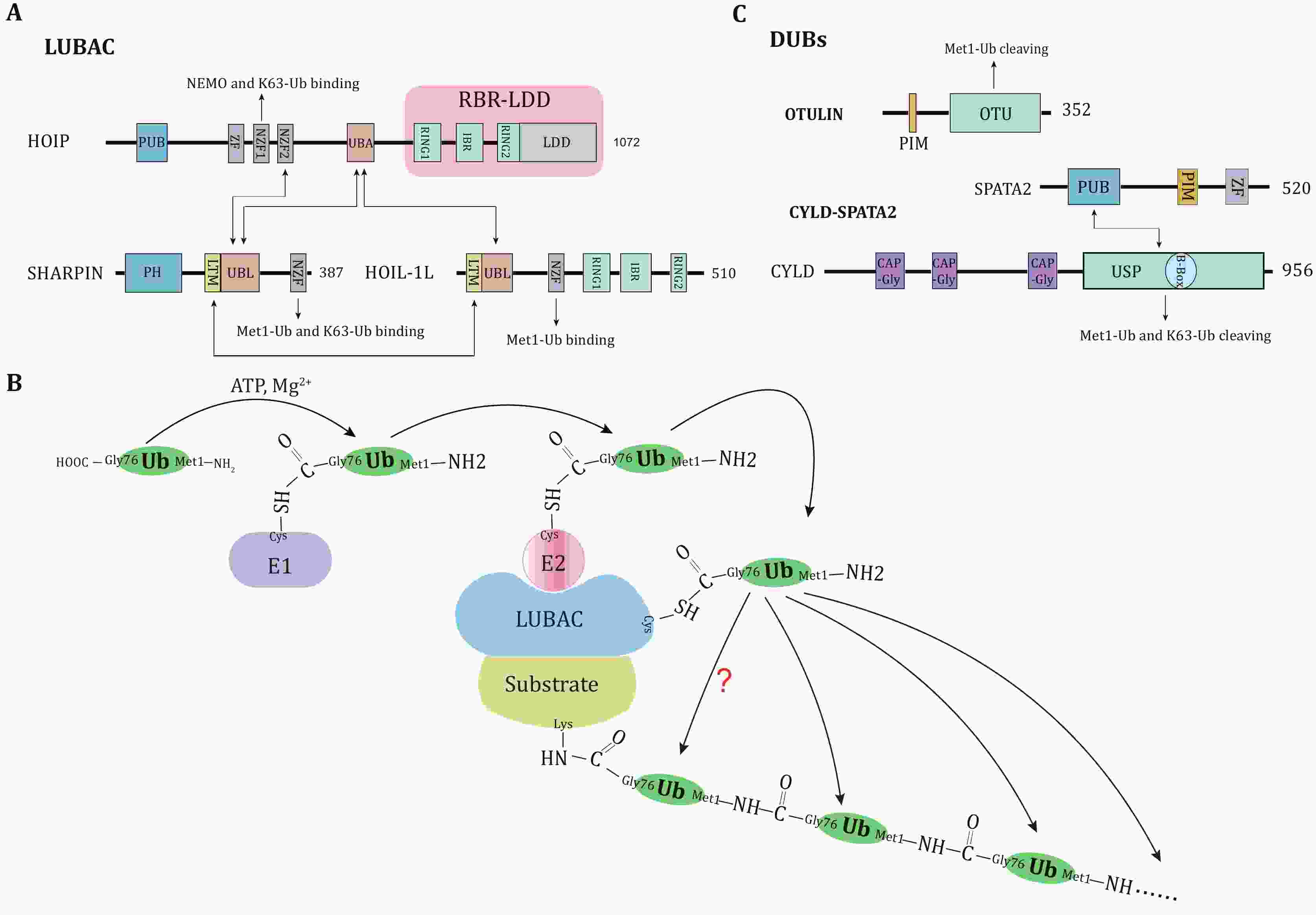



 Submit Your Paper
Submit Your Paper For Author
For Author Templates
Templates




 (86-10) 64888458
(86-10) 64888458





 E-mail alert
E-mail alert RSS
RSS